this post was submitted on 25 Oct 2024
350 points (97.0% liked)
Curated Tumblr
3944 readers
526 users here now
For preserving the least toxic and most culturally relevant Tumblr heritage posts.
Image descriptions and plain text captions of written content are expected of all screenshots. Here are some image text extractors (I looked these up quick and will gladly take FOSS recommendations):
-web
-iOS
Please begin copied raw text posts (lacking a screenshot that makes it apparent it is from Tumblr) with:
# This has been reposted here to Lemmy as part of the "Curated Tumblr Project."
I made the icon using multiple creative commons svg resources, the banner is this.
founded 1 year ago
MODERATORS
you are viewing a single comment's thread
view the rest of the comments
view the rest of the comments
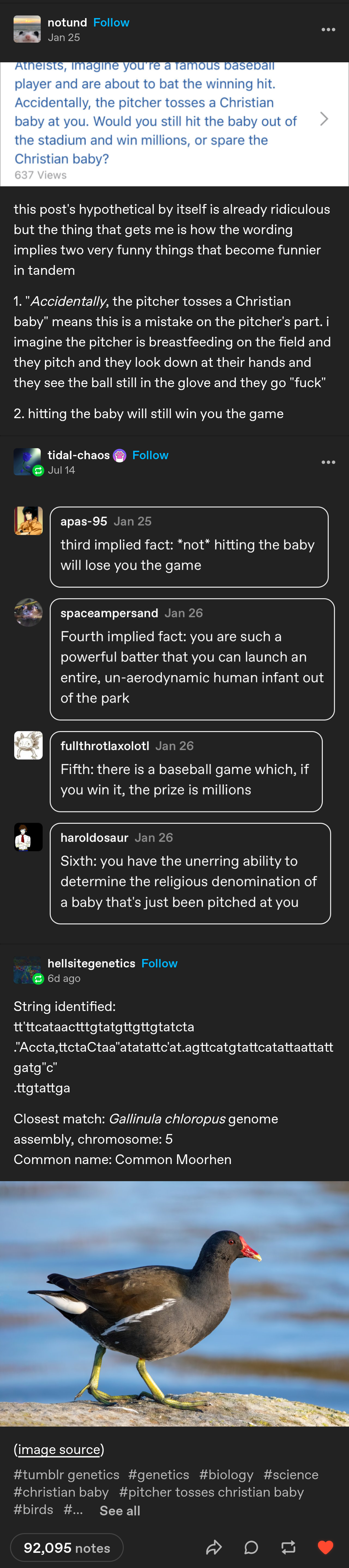
The genomes have likely been indexed to make finding results faster. Google doesn't search the entire internet when you make a query :P
I know that similar computational problems use indexing and vector-space representation but how would you build an index of TiBs of almost-random data that makes it faster to find the strictly closest match of an arbitrarily long sequence? I can think of some heuristics, such as bitmapping every occurrence of any 8-pair sequence across each kibibit in the list. A query search would then add the bitmaps of all 8-pair sequences within the query including ones with up to 2 errors, and using the resulting map to find "hotspots" to be checked with brute force. This will decrease the computation and storage access per query but drastically increase the storage size, which is already hard to manage.
However, efficient fuzzy string matching in giant datasets is an interesting problem that computer scientists must have encountered before. Can you find a good paper that works well with random, non-delimited data instead of just using the approach of word-based indices for human languages like Lucene and OpenFTS?
As per my other post, this person isn't doing any of that.
But, since you asked for papers on generic matching algorithms, I found this during the silent conniption fit you sent me into after suggesting that some random tumblr user plugged a tumblr bot directly into a state of the art genomics db.
https://link.springer.com/article/10.1007/s11227-022-04673-3
Please note that while, yes, they ran this test on a standard office computer, they were only searching against 12 million characters.
A single tebibyte of characters would be more like 1 trillion characters. A pebibyte would be more like 1 ~~quintillion~~ quadrillion.
... much, much, much longer processing times.
Edit: Used the wrong word for stupendously large numbers that start with q.
Yeah good point, not a trivial undertaking. I'm not an expert in that area but maybe elasticsearch or similar technology is able to find matches. Although I have no idea how that works under the hood